Function for prediction at new locations for single-species integrated occupancy models
predict.intPGOcc.RdThe function predict collects posterior predictive samples for a set of new locations given an object of class `intPGOcc`.
Usage
# S3 method for intPGOcc
predict(object, X.0, ...)Arguments
- object
an object of class intPGOcc
- X.0
the design matrix for prediction locations. This should include a column of 1s for the intercept. Covariates should have the same column names as those used when fitting the model with
intPGOcc.- ...
currently no additional arguments
Author
Jeffrey W. Doser doserjef@msu.edu,
Andrew O. Finley finleya@msu.edu
Value
An object of class predict.intPGOcc that is a list comprised of:
- psi.0.samples
a
codaobject of posterior predictive samples for the latent occurrence probability values.- z.0.samples
a
codaobject of posterior predictive samples for the latent occurrence values.
The return object will include additional objects used for standard extractor functions.
Examples
set.seed(1008)
# Simulate Data -----------------------------------------------------------
J.x <- 10
J.y <- 10
J.all <- J.x * J.y
# Number of data sources.
n.data <- 4
# Sites for each data source.
J.obs <- sample(ceiling(0.2 * J.all):ceiling(0.5 * J.all), n.data, replace = TRUE)
# Replicates for each data source.
n.rep <- list()
for (i in 1:n.data) {
n.rep[[i]] <- sample(1:4, size = J.obs[i], replace = TRUE)
}
# Occupancy covariates
beta <- c(0.5, 1)
p.occ <- length(beta)
# Detection covariates
alpha <- list()
for (i in 1:n.data) {
alpha[[i]] <- runif(2, -1, 1)
}
p.det.long <- sapply(alpha, length)
p.det <- sum(p.det.long)
# Simulate occupancy data.
dat <- simIntOcc(n.data = n.data, J.x = J.x, J.y = J.y, J.obs = J.obs,
n.rep = n.rep, beta = beta, alpha = alpha, sp = FALSE)
y <- dat$y
X <- dat$X.obs
X.p <- dat$X.p
sites <- dat$sites
# Package all data into a list
occ.covs <- X[, 2, drop = FALSE]
colnames(occ.covs) <- c('occ.cov')
det.covs <- list()
# Add covariates one by one
det.covs[[1]] <- list(det.cov.1.1 = X.p[[1]][, , 2])
det.covs[[2]] <- list(det.cov.2.1 = X.p[[2]][, , 2])
det.covs[[3]] <- list(det.cov.3.1 = X.p[[3]][, , 2])
det.covs[[4]] <- list(det.cov.4.1 = X.p[[4]][, , 2])
data.list <- list(y = y,
occ.covs = occ.covs,
det.covs = det.covs,
sites = sites)
J <- length(dat$z.obs)
# Initial values
inits.list <- list(alpha = list(0, 0, 0, 0),
beta = 0,
z = rep(1, J))
# Priors
prior.list <- list(beta.normal = list(mean = 0, var = 2.72),
alpha.normal = list(mean = list(0, 0, 0, 0),
var = list(2.72, 2.72, 2.72, 2.72)))
n.samples <- 5000
out <- intPGOcc(occ.formula = ~ occ.cov,
det.formula = list(f.1 = ~ det.cov.1.1,
f.2 = ~ det.cov.2.1,
f.3 = ~ det.cov.3.1,
f.4 = ~ det.cov.4.1),
data = data.list,
inits = inits.list,
n.samples = n.samples,
priors = prior.list,
n.omp.threads = 1,
verbose = TRUE,
n.report = 1000,
n.burn = 4000,
n.thin = 1)
#> ----------------------------------------
#> Preparing to run the model
#> ----------------------------------------
#> ----------------------------------------
#> Model description
#> ----------------------------------------
#> Integrated Occupancy Model with Polya-Gamma latent
#> variable fit with 81 sites.
#>
#> Integrating 4 occupancy data sets.
#>
#> Samples per Chain: 5000
#> Burn-in: 4000
#> Thinning Rate: 1
#> Number of Chains: 1
#> Total Posterior Samples: 1000
#>
#>
#> Source compiled with OpenMP support and model fit using 1 thread(s).
#>
#> ----------------------------------------
#> Chain 1
#> ----------------------------------------
#> Sampling ...
#> Sampled: 1000 of 5000, 20.00%
#> -------------------------------------------------
#> Sampled: 2000 of 5000, 40.00%
#> -------------------------------------------------
#> Sampled: 3000 of 5000, 60.00%
#> -------------------------------------------------
#> Sampled: 4000 of 5000, 80.00%
#> -------------------------------------------------
#> Sampled: 5000 of 5000, 100.00%
summary(out)
#>
#> Call:
#> intPGOcc(occ.formula = ~occ.cov, det.formula = list(f.1 = ~det.cov.1.1,
#> f.2 = ~det.cov.2.1, f.3 = ~det.cov.3.1, f.4 = ~det.cov.4.1),
#> data = data.list, inits = inits.list, priors = prior.list,
#> n.samples = n.samples, n.omp.threads = 1, verbose = TRUE,
#> n.report = 1000, n.burn = 4000, n.thin = 1)
#>
#> Samples per Chain: 5000
#> Burn-in: 4000
#> Thinning Rate: 1
#> Number of Chains: 1
#> Total Posterior Samples: 1000
#> Run Time (min): 0.0064
#>
#> Occurrence (logit scale):
#> Mean SD 2.5% 50% 97.5% Rhat ESS
#> (Intercept) 0.9305 0.3615 0.2652 0.9076 1.7166 NA 299
#> occ.cov 1.2033 0.3474 0.5949 1.1794 1.9201 NA 329
#>
#> Data source 1 Detection (logit scale):
#> Mean SD 2.5% 50% 97.5% Rhat ESS
#> (Intercept) -0.4199 0.3375 -1.0725 -0.4245 0.2597 NA 685
#> det.cov.1.1 -0.6578 0.4124 -1.5288 -0.6281 0.0707 NA 471
#>
#> Data source 2 Detection (logit scale):
#> Mean SD 2.5% 50% 97.5% Rhat ESS
#> (Intercept) -0.9276 0.3966 -1.7125 -0.9096 -0.1879 NA 597
#> det.cov.2.1 0.9189 0.4664 0.0876 0.9001 1.8753 NA 543
#>
#> Data source 3 Detection (logit scale):
#> Mean SD 2.5% 50% 97.5% Rhat ESS
#> (Intercept) 0.1401 0.2675 -0.3877 0.1387 0.6817 NA 667
#> det.cov.3.1 -1.0872 0.3194 -1.7360 -1.0744 -0.5203 NA 450
#>
#> Data source 4 Detection (logit scale):
#> Mean SD 2.5% 50% 97.5% Rhat ESS
#> (Intercept) -0.0591 0.3225 -0.6712 -0.0555 0.5537 NA 548
#> det.cov.4.1 0.9524 0.4511 0.1711 0.9159 1.8861 NA 609
#>
# Prediction
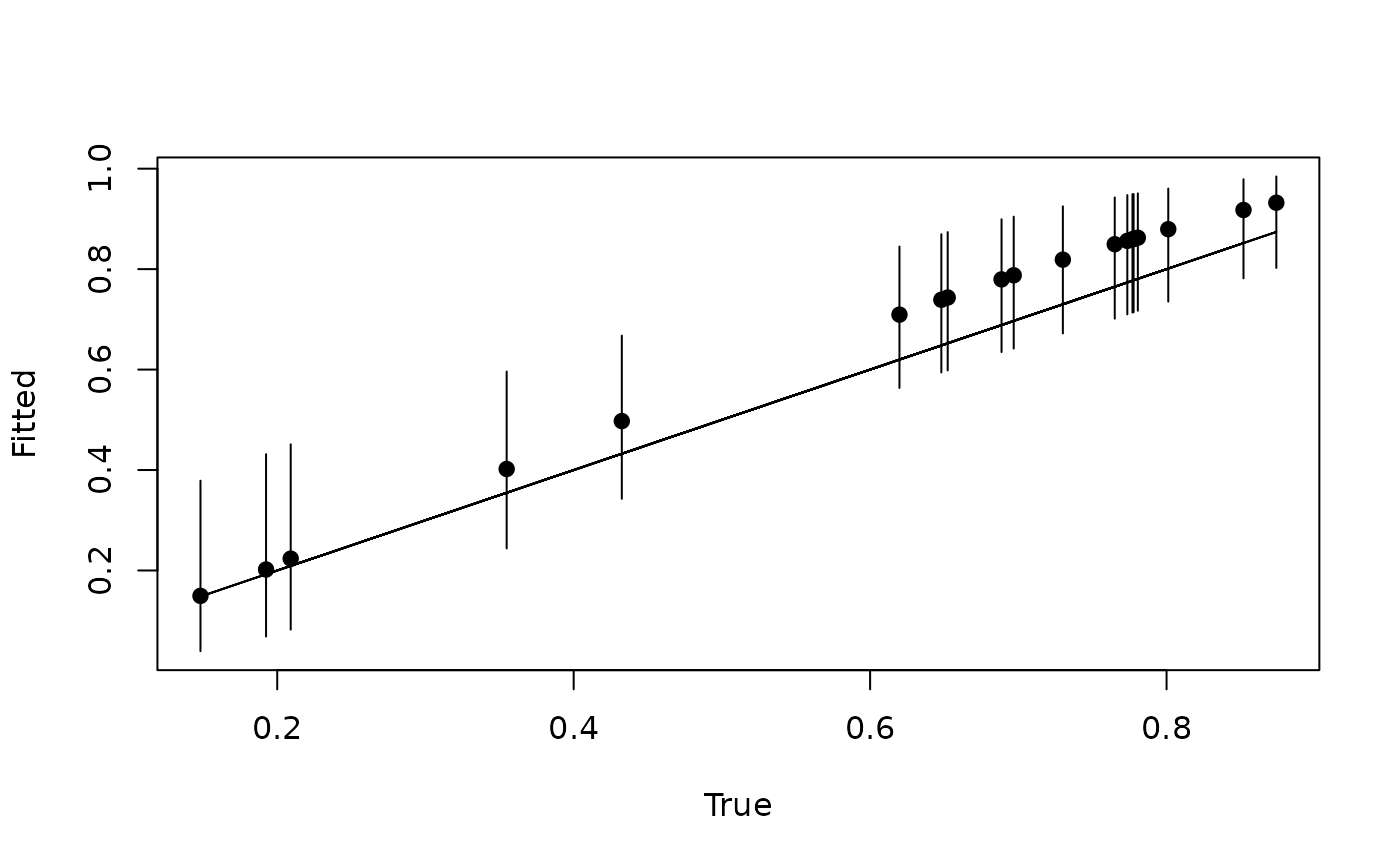
X.0 <- dat$X.pred
psi.0 <- dat$psi.pred
out.pred <- predict(out, X.0)
psi.hat.quants <- apply(out.pred$psi.0.samples, 2, quantile, c(0.025, 0.5, 0.975))
plot(psi.0, psi.hat.quants[2, ], pch = 19, xlab = 'True',
ylab = 'Fitted', ylim = c(min(psi.hat.quants), max(psi.hat.quants)))
segments(psi.0, psi.hat.quants[1, ], psi.0, psi.hat.quants[3, ])
lines(psi.0, psi.0)